High-resolution natural killer cell phenotyping by mass cytometry in pediatric transplant recipients
Wenming Zhang1, Josselyn K. Pena1,2, Potchara Boonrat1,2, James T. Harden1,2, Carlos O. Esquivel1, Olivia M. Martinez1,2, Sheri M. Krams1,2.
1Department of Surgery, Stanford University School of Medicine, Stanford, CA, United States; 2Stanford Immunology, Stanford University School of Medicine, Stanford, CA, United States
Introduction: Solid organ transplant remains the best and often only treatment for children with end-stage diseases. Despite recent advances in immunosuppressive agents, complications caused by viral infections continue to be a major barrier to graft and patient survival. Epstein-Barr virus (EBV), a broadly disseminated human gamma herpes virus, is associated with devastating complications due to its oncogenicity. In transplant recipients, post-transplant lymphoproliferative disorder (PTLD) is the most extreme manifestation of defective immune control of EBV infection. Natural Killer (NK) cells are lymphocytes of the innate immune system that can rapidly respond to virally infected cells via cytotoxic pathways or production of cytokines. While it is known that NK cells are important in the control of EBV infection, the specific populations involved remain to be determined. Previous work in our laboratory has shown that NKG2A+ NK cells are enriched in the NK cell population that can recognize and respond to B cells latently infected with EBV. The purpose of this study was to: 1) capture potential signatures of NK cell response to latent EBV infection in vivo and 2) achieve high-dimensional phenotyping of NK cell populations associated with EBV.
Method: We established a custom 48-marker mass cytometry panel of phenotypic and functional NK cell markers, including CD56, CD16, NKG2A, NKG2C, NKG2C, NKp46, CD57, HLA-DR, KIRs, TIGIT, Siglec-7, and others. Using mass cytometry, we analyzed NK cell subsets in pediatric transplant patients. Peripheral blood NK cells (3-5x106 PBMCs) were obtained 6-9 months post-transplant from 33 participants who were either EBV naïve (n=21) or EBV seropositive (n=12). Normalized cytometry data were analyzed in Cellengine. FlowSOM clustering was conducted in R.
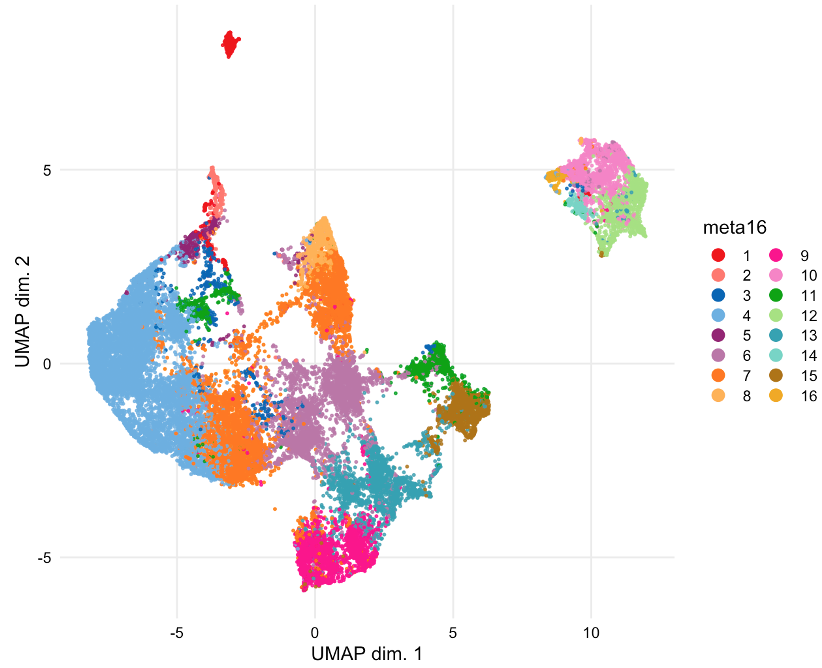
Results: Manual gating was used to identify NK cells (CD3- CD14- CD19- CD56+) within total live cells (Cisplatinlo) in patient PBMCs. 16 unique CD56+ cell clusters were identified by FlowSOM. Cells were sampled for nonlinear dimensionality reduction using uniform manifold approximation and projection (UMAP). The identified NK cell subsets are visualized by UMAP (Figure 1) and UMAP maps are colored according to the arcsinh-transformed expression level of CD56, CD16, NKp46, NKG2A, NKG2C, and CD57 (Figure 2). Our findings indicate NK cells include subsets that are phenotypically and functionally diverse in EBV+ and EBV- individuals which differentially express NK cell activation and inhibitory receptors, including NKG2A, NKG2C, and NKp46.
Conclusion: Ongoing analysis to identify NK cell subsets that control EBV infection will potentially lead to improved therapies for transplant recipients.

References:
[1] Hatton, Olivia et al. “NKG2A-Expressing Natural Killer Cells Dominate the Response to Autologous Lymphoblastoid Cells Infected with Epstein-Barr Virus.” Frontiers in Immunology vol. 7 607. 15 Dec. 2016
[2] Mbiribindi, Berenice et al. “Epstein-Barr virus peptides derived from latent cycle proteins alter NKG2A + NK cell effector function.” Scientific Reports vol. 10,1 19973. 17 Nov. 2020
Lectures by Sheri M. Krams
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Tue-28 13:00 - 14:00 |
Closing plenary and presentation highlight of basic science and clinical science abstracts | Highlight of Basic Science Abstracts | Zilker 3-4 |
|
Mon-27 10:00 - 11:00 |
Basic Science / HLA | High-dimensional profiling of pediatric immune responses to solid organ transplantation | Hill Country CD |
|
Mon-27 10:00 - 11:00 |
Basic Science / HLA | High-resolution natural killer cell phenotyping by mass cytometry in pediatric transplant recipients | Hill Country CD |
|
Sun-26 15:00 - 16:00 |
IPTA-ASHI: New opportunities in HLA | CyTOF for immune profiling of pediatric transplant recipients | Zilker 3-4 |
